Other MISO Tools
The following tools can help you with planning or finding things within MISO. They can be found by clicking the appropriate link in the Tools list in the menu on the left side of the screen.
Index Distance
When you run multiple libraries in a sequencing lane or partition, known as multiplexing, indices are used to identify the individual libraries and demultiplex (separate) the data afterwards. If two or more libraries in the pool have identical or near-identical indices, demultiplexing may be more difficult or even impossible. The Index Distance tool can help you identify indices which may be inadvisable to multiplex together.
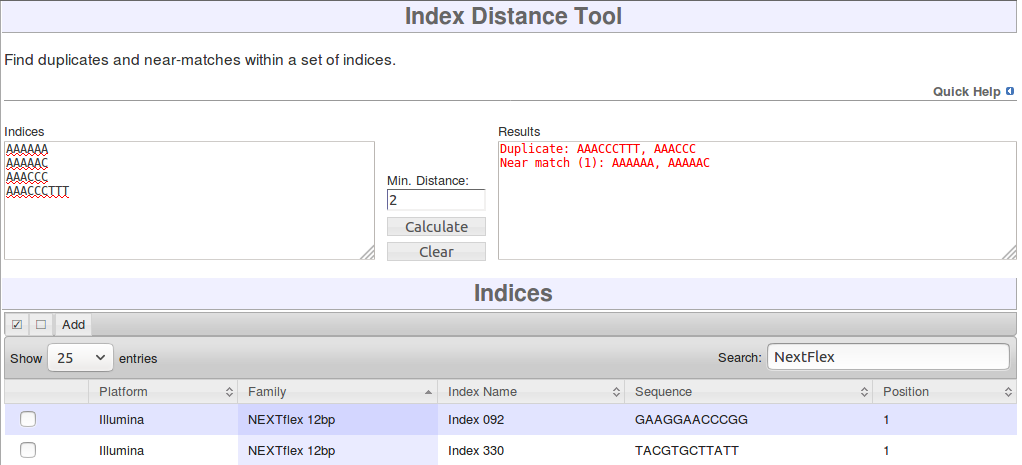 Index Distance tool
Index Distance tool
To use the tool, enter all of the indices you wish to compare into the Indices box. You can use the Indices list below to search for indices and then select and add them, or you can type the index sequences directly into the Indices box. For dual barcodes, enter both indices together as if they were one. e.g. 'AAAAAA' index 1 and 'CCCCCC' index 2 should be entered as 'AAAAAACCCCCC'.
In the Min. Mismatches box, you can choose the minimum number of base mismatches that the sequences must have. For example, 'AAAAAA' and 'AAAACC' have 2 mismatches. You will be warned of any sequences which have a lower number of mismatches than that entered here. If indices are of different lengths, only the length of the shortest one will be considered, and the extra bases on the longer index will be ignored. For example, 'AAAAAA' and 'AAAAAACC' will be considered duplicates.
When you have made your selections, click the Calculate button and the Results box will be updated to show any duplicates and near duplicates.
Another version of this tool can be found within this documentation here. This version requires that you enter index sequences yourself, as it is not connected to a MISO database from which it can look up indices. It is available to people who don't have access to MISO, however.
Index Search
If you have a list of index sequences, but do not know either the name of the index family containing them, or whether they exist in MISO at all, you can use the Index Search tool to check.
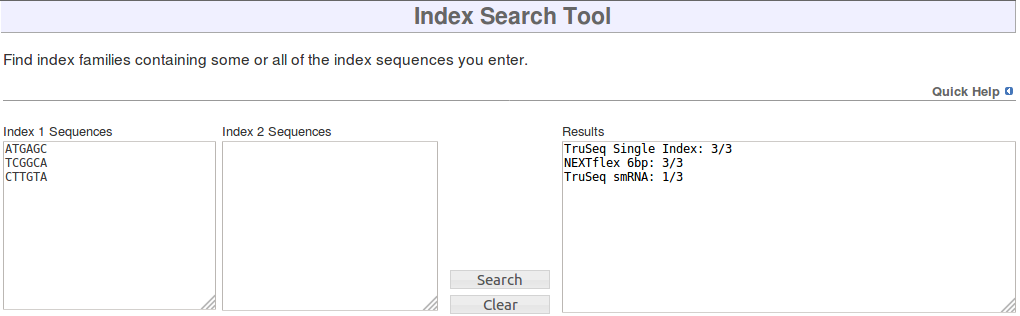 Index Search tool
Index Search tool
To use the tool, enter the index sequences you are looking for into the Index 1 Sequences and Index 2 Sequences boxes. The Index 2 Sequences box should be left empty if you are not looking for dual indices. Click the Search button and the Results box will be updated to show every index family containing any of the index sequences you entered, along with how many of the indices each family contains.
Identity Search
Note: This tool only appears if your site uses detailed sample mode.
The Identity Search tool can be used to find identity (donor/organism) samples with a specified external name, and to see all of the samples descended from the resulting identities. This is useful if you're not sure whether you've received samples from this donor or organism before, or if you want to see all of the samples related to it.
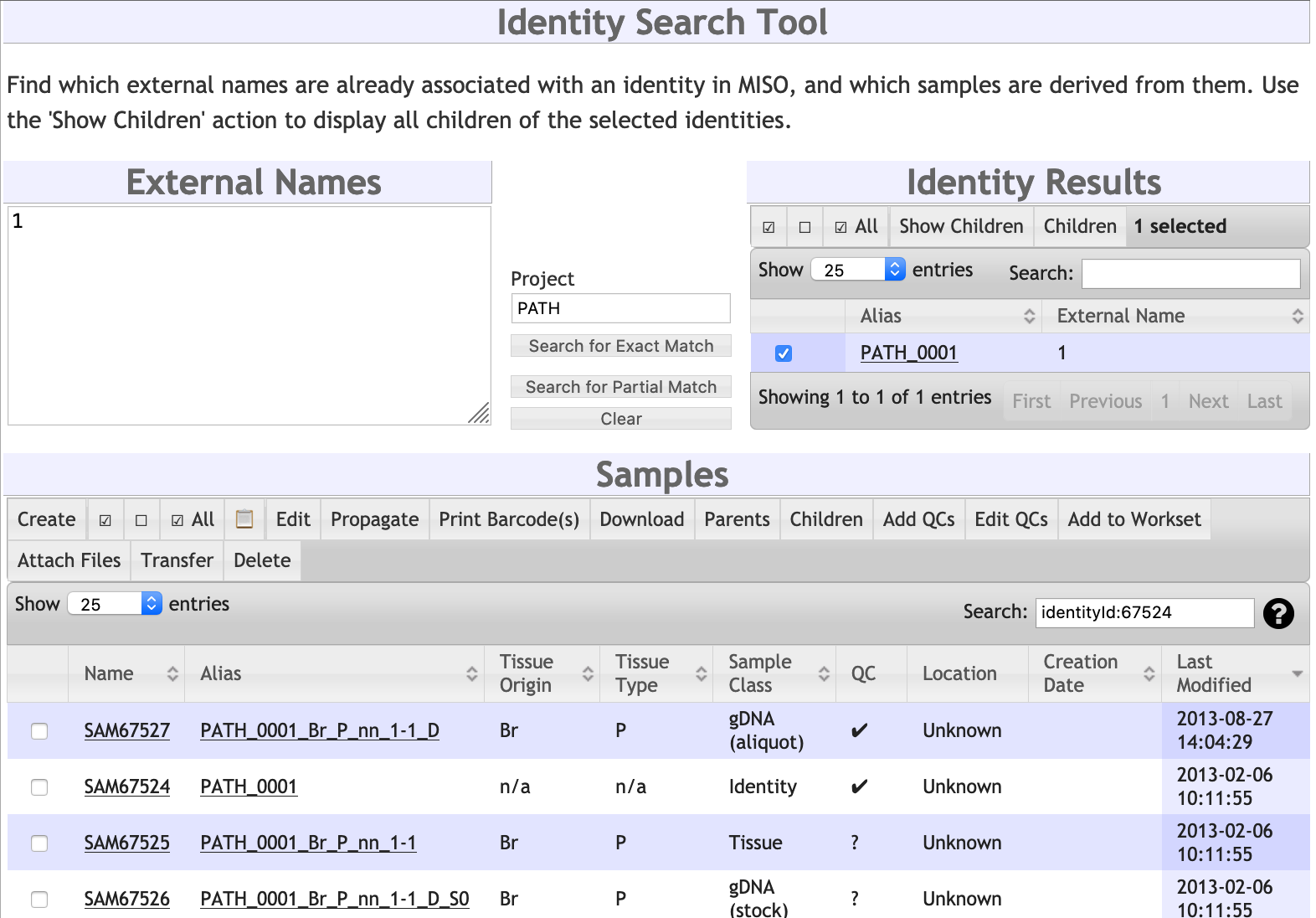 Identity Search tool
Identity Search tool
To use the Identity Search, enter the external names you're looking for in the External Names box. Optionally, you can enter the short name of the project you wish to search in.
After clicking one of the Search buttons -- either for exact or partial matches -- matching identity samples will be listed in the Identity Results box. To view all children of one or more of these identities, select the identities and click the 'Show Children' button. The children will be displayed in the Samples list below.
The Identity Results list also has a 'Children' button, which is useful if you want to perform bulk actions on a particular type of descendant, such as tissues, or libraries.